PowQ
A graphical tool for evaluating the power of variance component multipoint linkage analysis studies through simulations.
PowQ
allows power calculations of variance component linkage analysis studies
through simulations for study samples that can comprise assorted family
structures, including extended pedigrees.
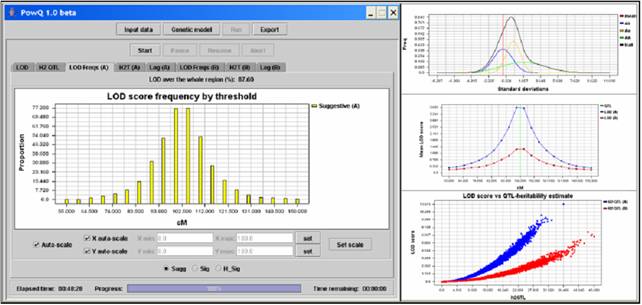
The
multipoint framework implemented in PowQ provides both accurate estimates of
the expected power and confidence intervals for the location of the QTL. When
the power study is evaluated in a region smaller than the expected confidence
interval for the QTL location (or in a single-point framework) it is likely to
provide an underestimate of the real dataset power.
PowQ
also allows power comparisons of nested samples, which are easily
calculated providing an initial sample and its possible extensions/reductions,
both in terms of family units and in terms of family members. Relative power
computation allows identification of the most informative and/or economical
sample.
The user selects one pedigree
file, or two for a comparative study, a map file with the markers position and
the QTL position, and a genetic model for the simulated trait.
Founder individuals of each
family belonging to the pedigree file are assumed to be unrelated, and founder
haplotypes are assumed to be in linkage equilibrium.
Power is computed by
randomly sampling from the inheritance space, assuming complete knowledge of
IBD sharing among pedigree members and counting the number of times the test
statistic falls above the required thresholds.
Credits
PowQ
was developed by Mario Falchi at the Twin
Research Unit and by Cesare Cappio
Borlino at Shardna
The variance component
linkage analysis approach follows the Merlin implementation (Abecasis et al
2001).
For
any comments and suggestions, please email: mario.falchi@kcl.ac.uk
Citation
PowQ can be cited with the following paper:
Falchi M, Borlino CC. PowQ:
a user-friendly package for the design of variance component multipoint linkage
analysis studies.
Bioinformatics. 2006 Jun 1;22(11):1404-5.